Research
Credit: Rachel Torrez/the Animation Lab
The Dassama Research Group performs research at the interface of chemistry and biology, relying on tools of chemistry to probe and explain biological phenomena. Two overarching questions drive our research program:
1) As drug resistant pathogenic bacterial strains arise, can we rationally develop/engineer new classes of antimicrobial agents designed to exploit key metabolic vulnerabilities in human pathogens?
2) Are there generalizable ways to modulate disease-relevant human proteins that are thus far intractable to small molecule drugs?
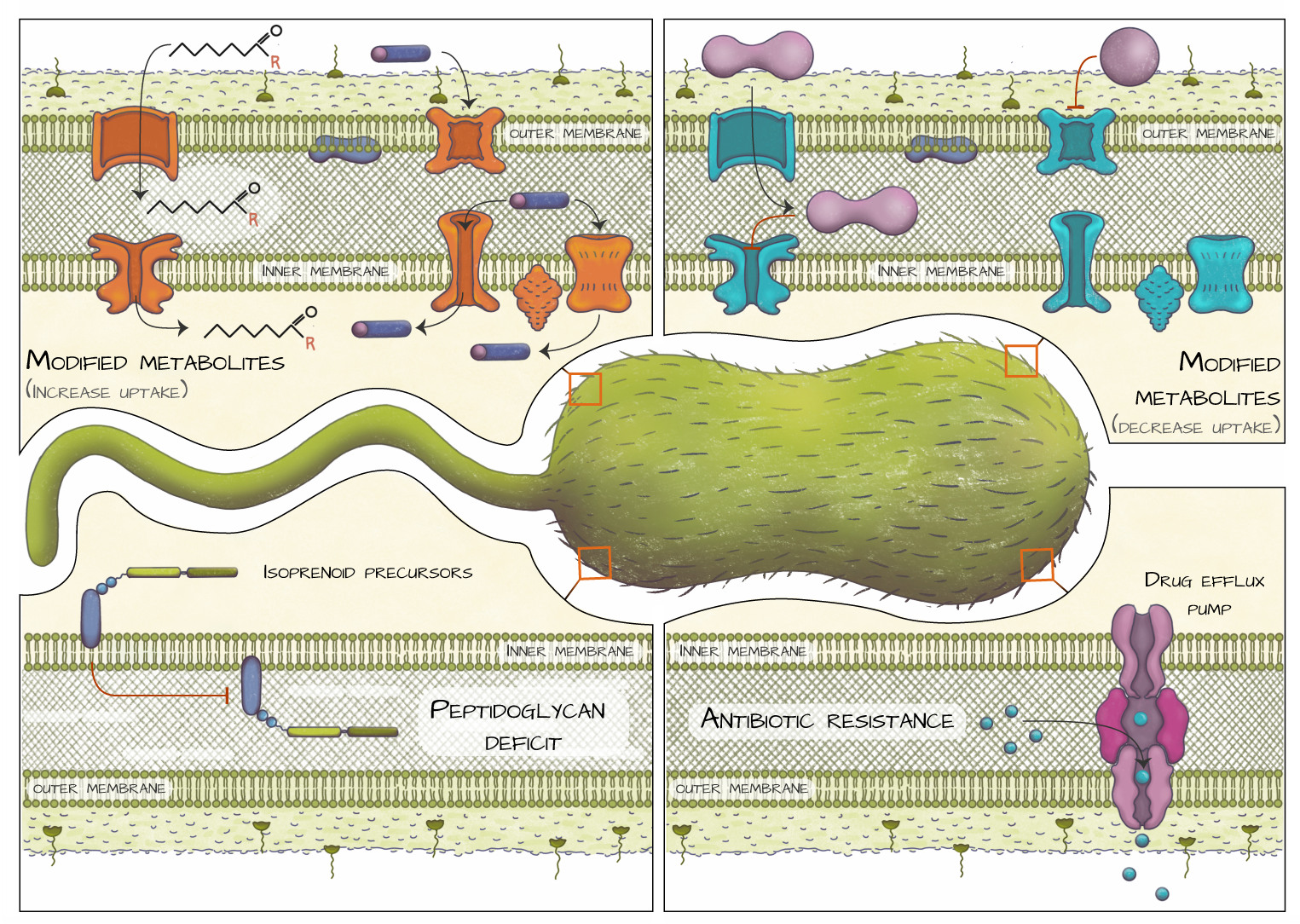
Bacterial metabolites
Credit: DrawInScience
Even in a relatively simple bacterium, metabolites must exist in specific locations to function properly: molecules involved in nutrient acquisition are secreted and subsequently taken up, lipids must travel to cell membranes, etc. Generally, transporter proteins mediate the trafficking of these metabolites, but at times there is poor understanding of how they function. Our research leverages bioinformatics, biochemistry, and biophysics to discover and characterize proteins that mediate the transport of metabolites. We focus on the transport of primary metabolites like lipids, which are important for cell viability, proliferation, and pathogenesis.
We also study the transport of secondary metabolites and antibiotics because they are relevant to multidrug resistance mediation efforts. In this realm, our work centers around integral membrane proteins called multidrug and toxic compound efflux (MATE) pumps that have emerged as key players in MDR, as their presence enables microbes to secrete multiple antibiotics. Our research efforts are focused on determining the substrate scope and transport mechanisms for these “divergent” MATE proteins.
Finally, we have substantial research efforts dedicated to mining the biosynthetic pathways to primary metabolites to aid the discovery of novel therapeutic targets.
Targeted modulation of proteins
Credit: DrawInScience
A significant fraction of the human proteome remains “undruggable” because it comprises proteins not amenable to inhibition by small molecules. In recent years, proximity-based approaches have been used to modulate some of these proteins, primarily through degradation with proteolysis targeting chimeras (PROTACs) and molecular glues. PROTACs were first reported two decades ago and promised as powerful tools for biological inquiry and for advancing human health. However, many of the proximity-based protein modulation strategies are incompatible with proteins that lack defined small molecule binding pockets. Our research focuses on these so-called “undruggable” proteins and employs protein-based ligands for their targeted modulation via the addition (or removal) of post-translational modifications (PTMs) that finely tune protein function. We discover ligands selective for challenging proteins (e.g., intrinsically disordered proteins) and functionalize these ligands to install or remove specific PTMs in a precise manner, and design strategies to deliver them to the relevant cells and tissues.